Biomolecular Control Systems
 |
Over several decades, control theory has significantly contributed to the design of control systems for engineered machines, influencing the evolution of major engineering disciplines like electrical, mechanical, and chemical engineering. In these fields, designing controllers involves creating electrical circuits or mechanical systems that incorporate components such as resistors, capacitors, inductors, springs, and dampers. More recently, control theory has made its way into molecular biology, particularly within the domains of systems and synthetic biology . It is anticipated that control theory will play a similar role in shaping these emerging fields, just as it has in more traditional engineering and scientific disciplines.
Although control theory is a mature field, the complex nature of biomolecular systems has introduced new challenges that require innovative control-theoretic tools . These tools must be tailored to the intricate interactions between biochemical molecules. Designing and implementing an effective control system in this context involves two key challenges. The first challenge is to develop a suitable control law that adheres to a specific architecture. Unlike in digital systems, where implementing a linear PID controller for instance is trivial, biomolecular systems demand controllers that conform to the structure of chemical reaction networks involving positivity constraints, fundamentally nonlinear interactions, and stochastic dynamics . As such the design process becomes significantly more restrictive and complex. The second challenge is ensuring that the controller can be biologically implemented . This involves identifying and integrating the appropriate biological components that will dynamically interact according to the control law. This challenge bridges the gap between theoretical mathematics and practical biology, which is not typically an issue in digital control systems implemented on computers.In biomolecular applications, both challenges are equally critical. Controllers must be specifically structured to be biologically viable and to ensure robust features like disturbance rejection or perfect adaptation. This requires a careful balance of mathematical modeling and practical biological implementation .
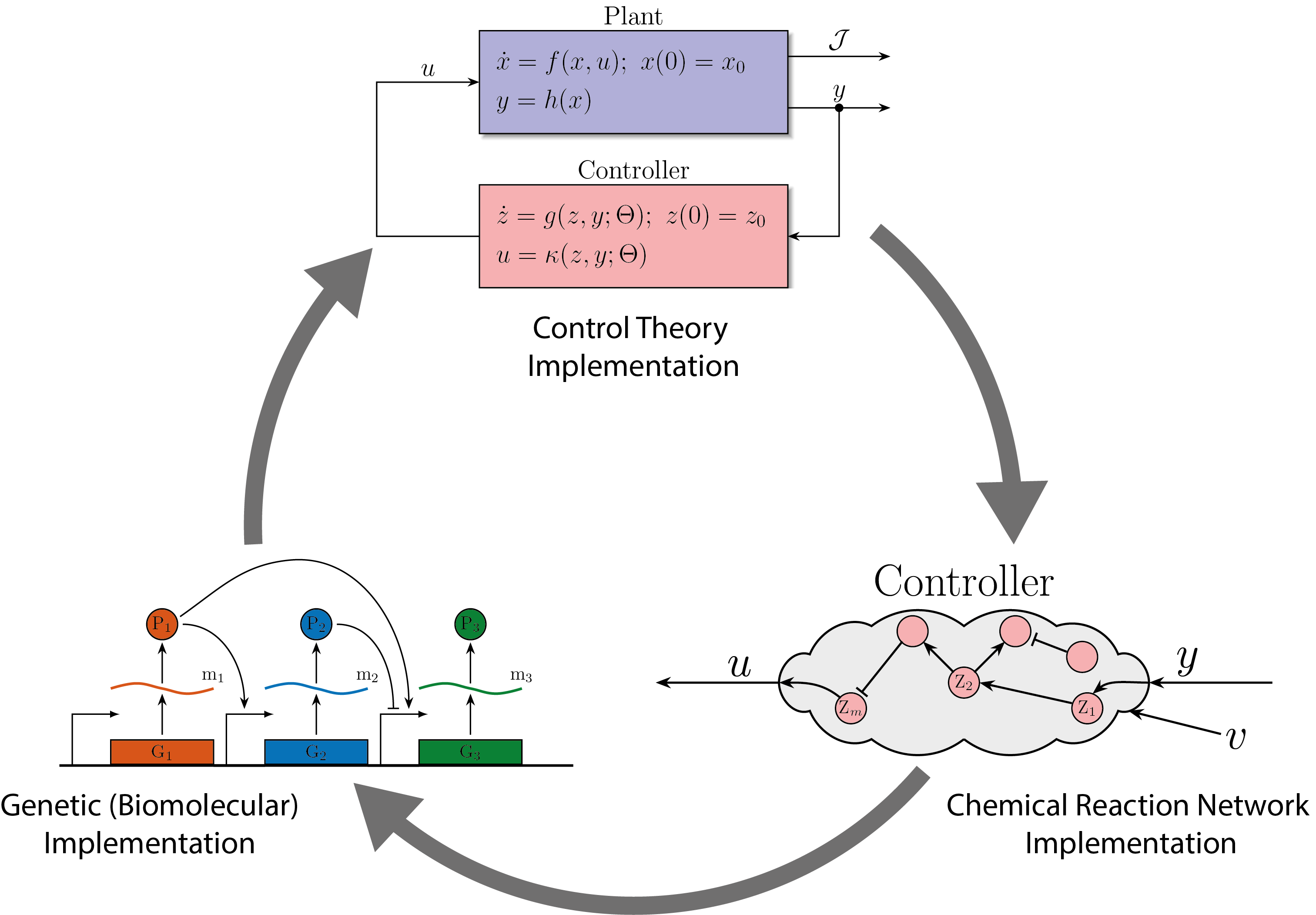
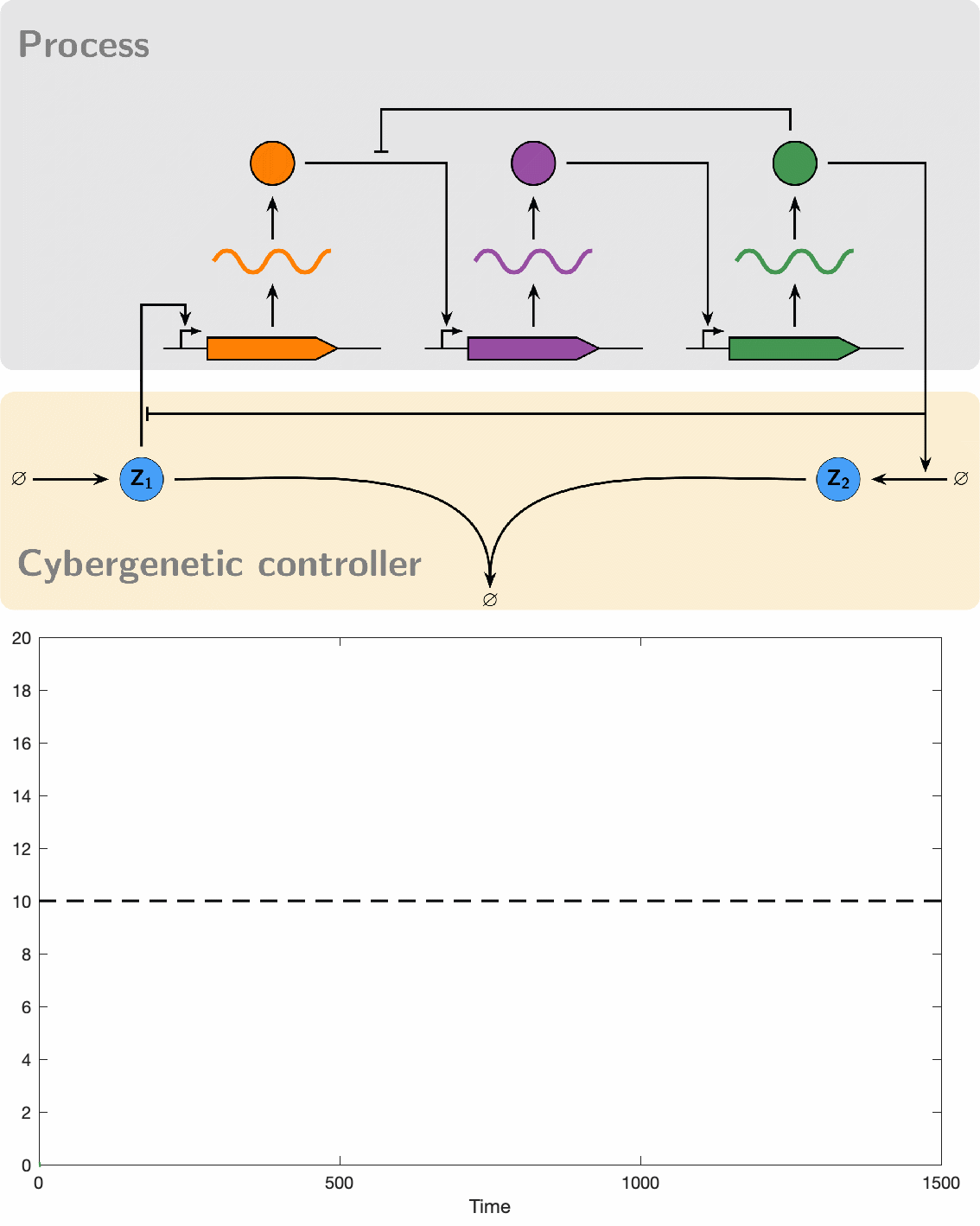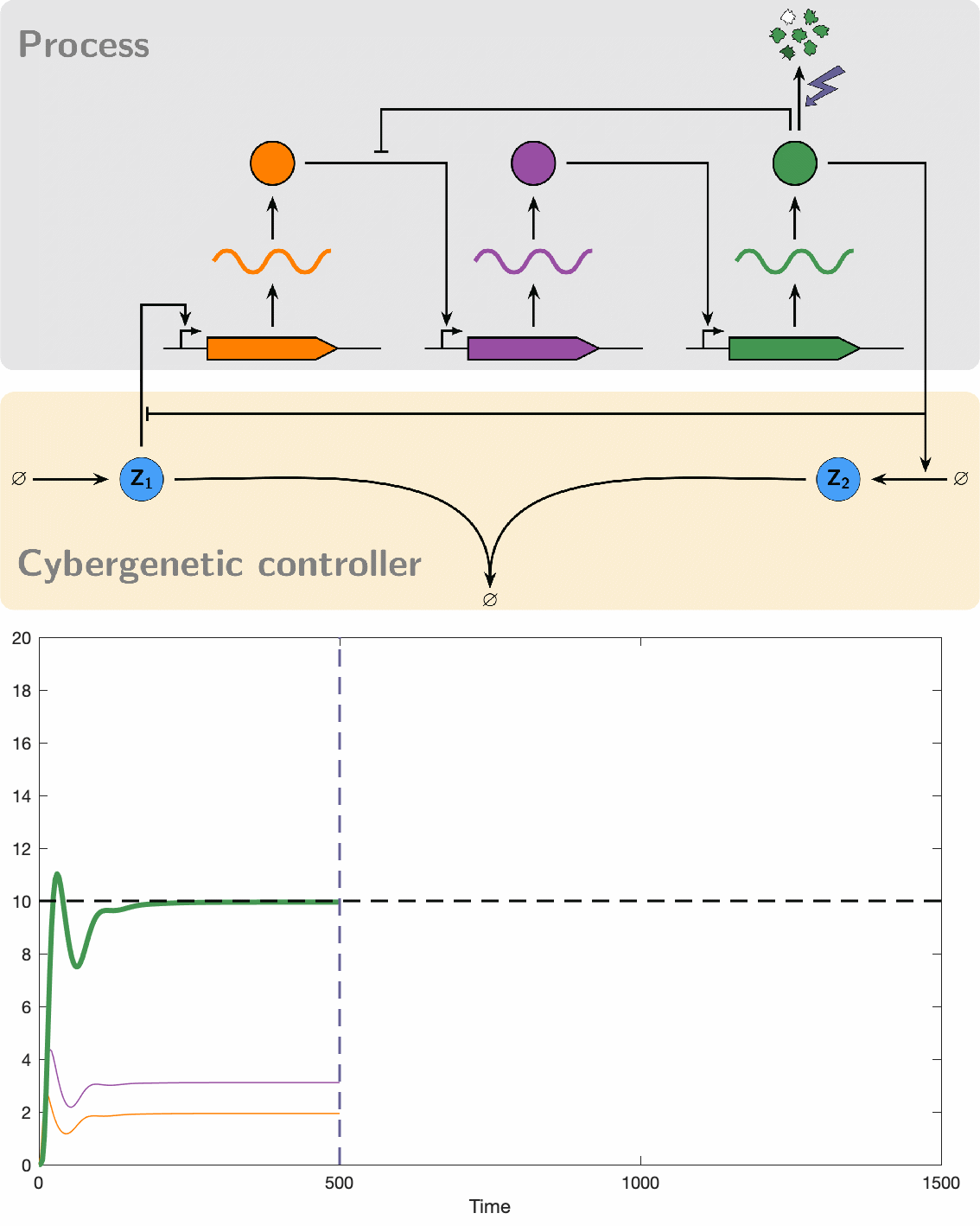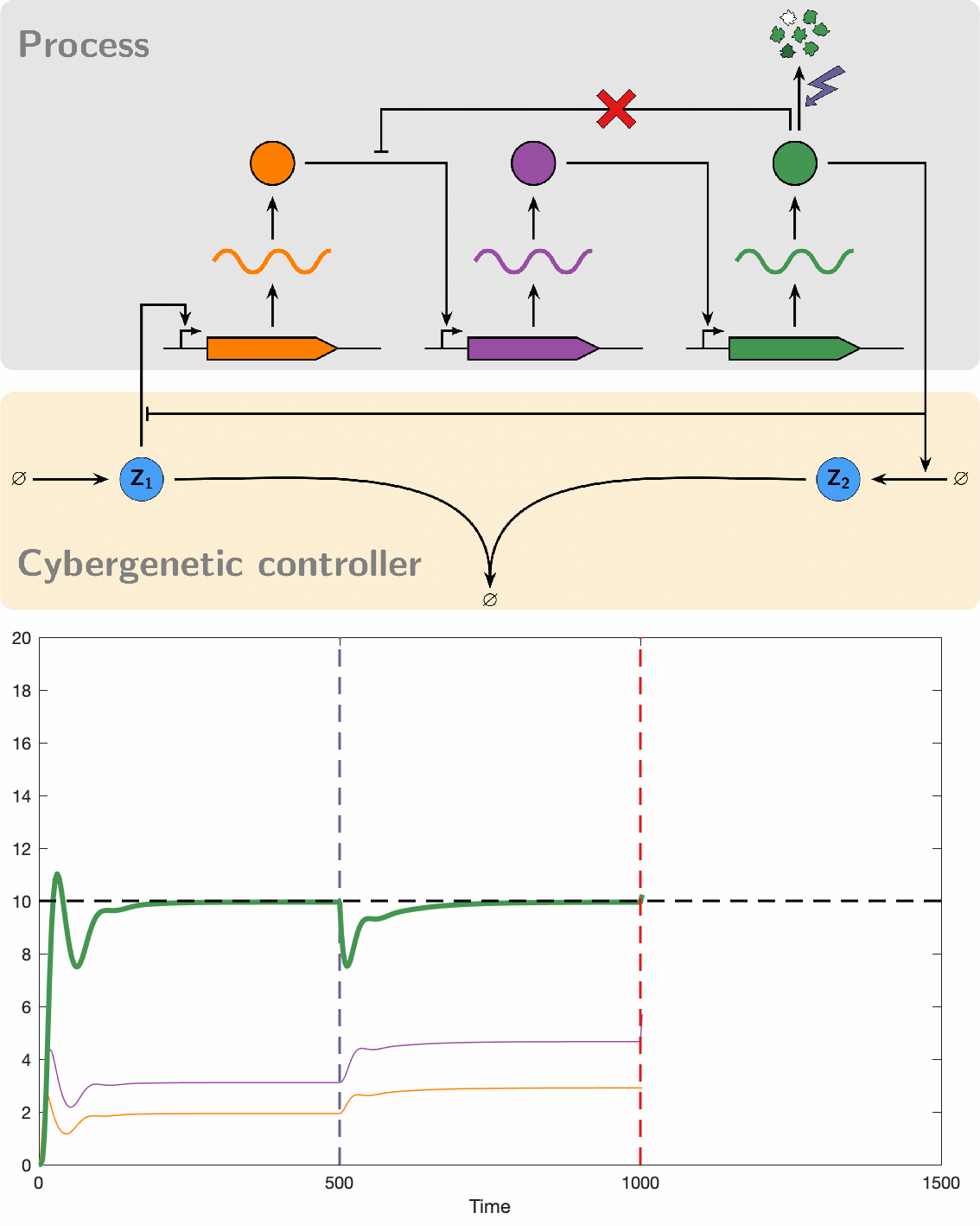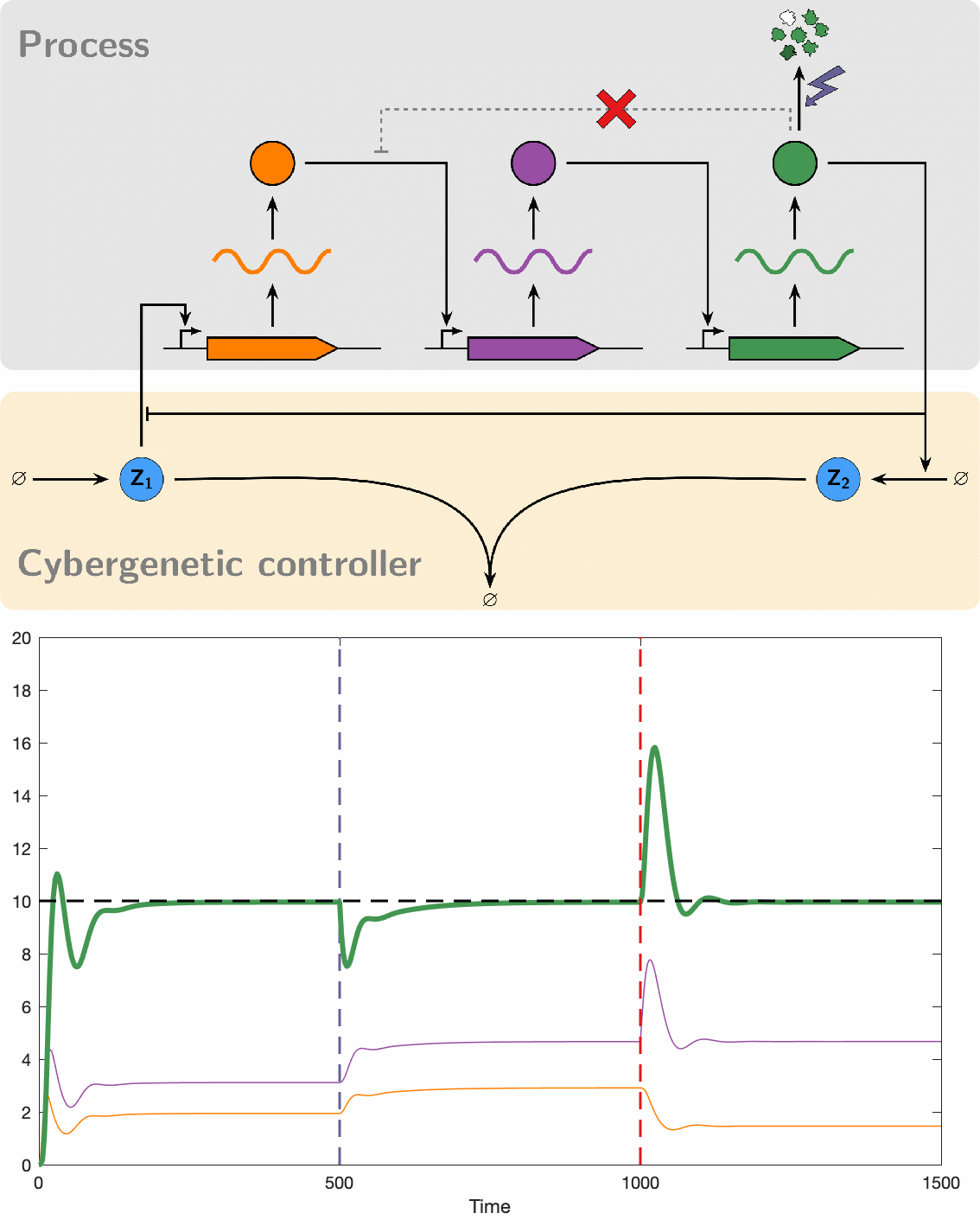
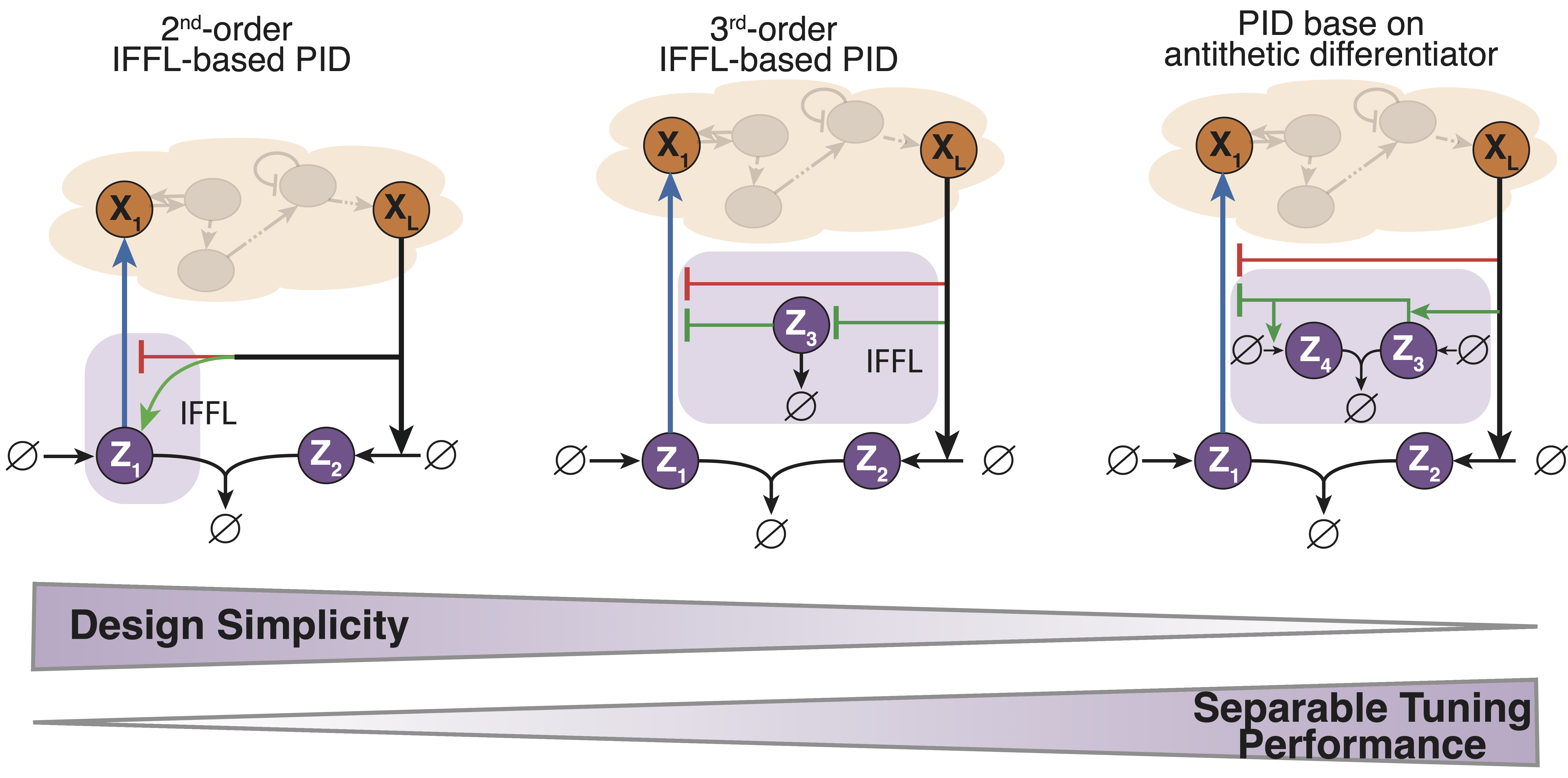
PID controllers are among the simplest and most widely used controllers for engineered machines. However, implementing PID controllers through chemical reaction networks presents significant challenges. Our objective was to systematically and rationally design a series of such controllers of increasing complexity. This approach allowed us to establish a hierarchy of biomolecular PID controllers, offering a tradeoff between simplicity in the design and separability of the PID components that makes the tuning a little easier and offer a better performance.
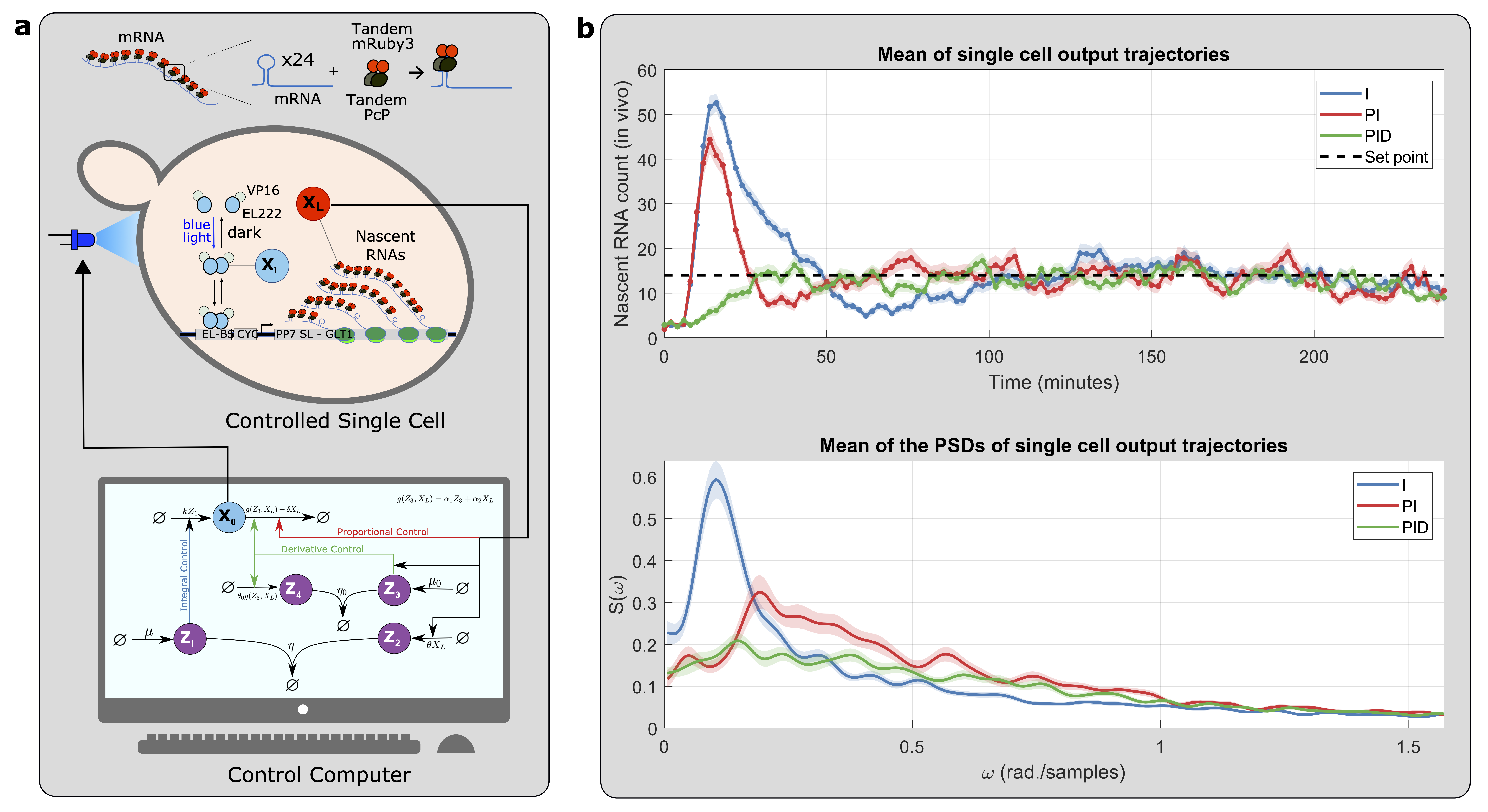
Our initial foray into experimenting with biomolecular controllers involved testing them on the cyberloop, a hybrid platform that integrates real biological circuits within cells (in vivo) under a microscope with stochastic computer simulations (in silico) of controllers. This is achieved through light stimulation and fluorescence measurement, allowing individual and simultaneous observation of multiple cells. Our experiments successfully demonstrated that these controllers can robustly maintain a desired setpoint, achieve high dynamic performance, and reduce noise, which often appears as cell-to-cell variability.
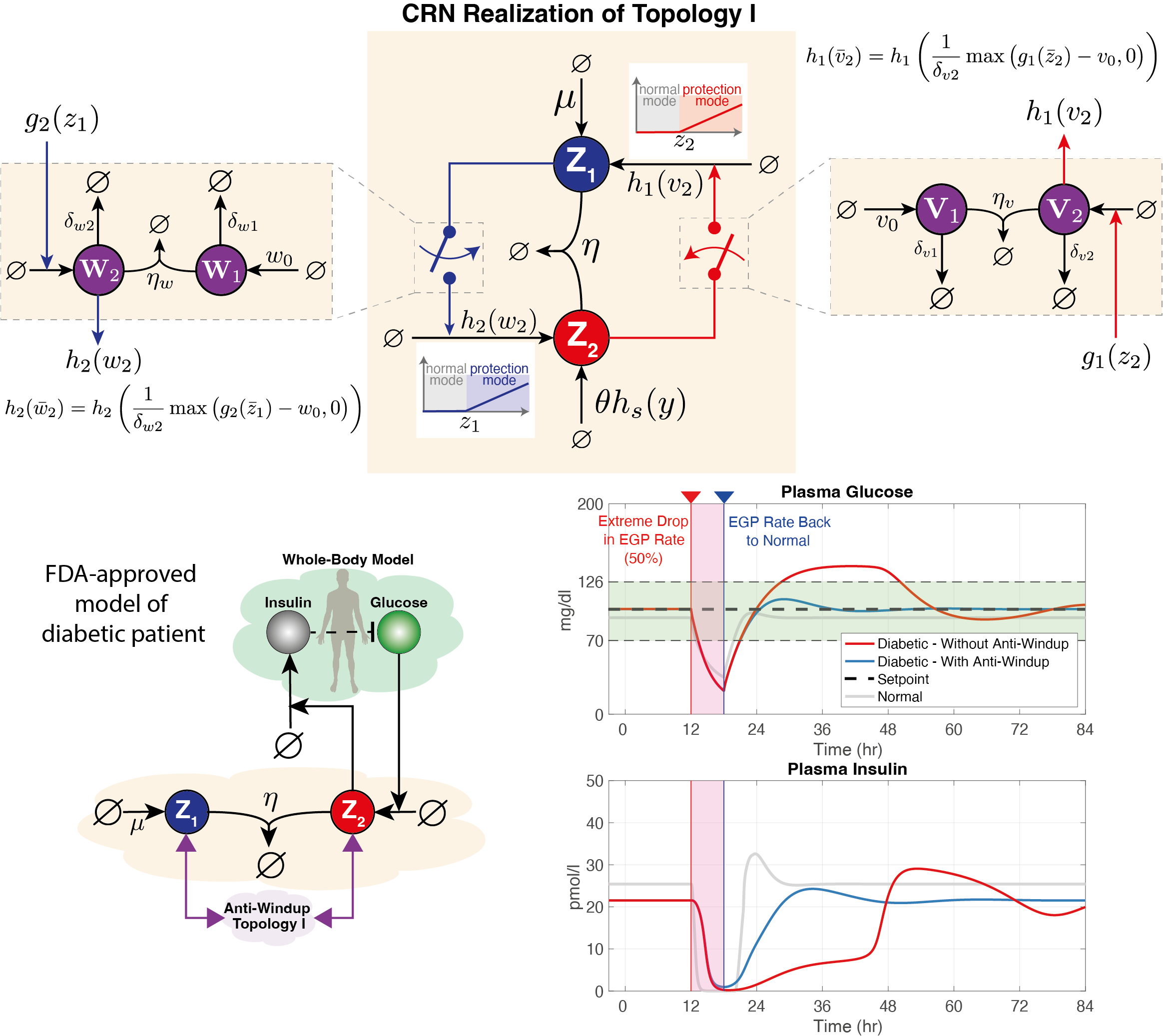
Despite their great promise, the effectiveness of biomolecular PID controllers effectiveness is hindered by several challenges, particularly integral windup. This issue occurs when the integral component of a controller accumulates errors over time, leading to significant overshooting or undershooting of the target setpoint. This is exacerbated in biomolecular systems due to common factors like saturation of molecular concentrations, promoter saturation, and limitations in resources such as enzyme concentrations and energy molecules. Such conditions can cause prolonged poor performance and even system instability. Integral windup can be thought of as a "trauma" to a system caused by a severe disturbance, with its effects persisting for a considerable time even after the disturbance has ceased. In this analogy, implementing an anti-windup strategy acts like a therapeutic intervention, allowing the controller to recover by effectively "forgetting" the disruptive event. We have developed various anti-windup topologies using chemical reaction networks and have also proposed genetic implementations. Additionally, we formulated a theorem that facilitates the analysis of anti-windup dynamics, connecting them to established strategies in control theory.
This search for genetic parts echoes the historical quest in electronics engineering that culminated in the transition from vacuum tubes to transistors. In our early efforts, we successfully utilized the hybridization properties of sense/anti-sense RNA to model and implement the first biomolecular PI controller in mammalian cells.
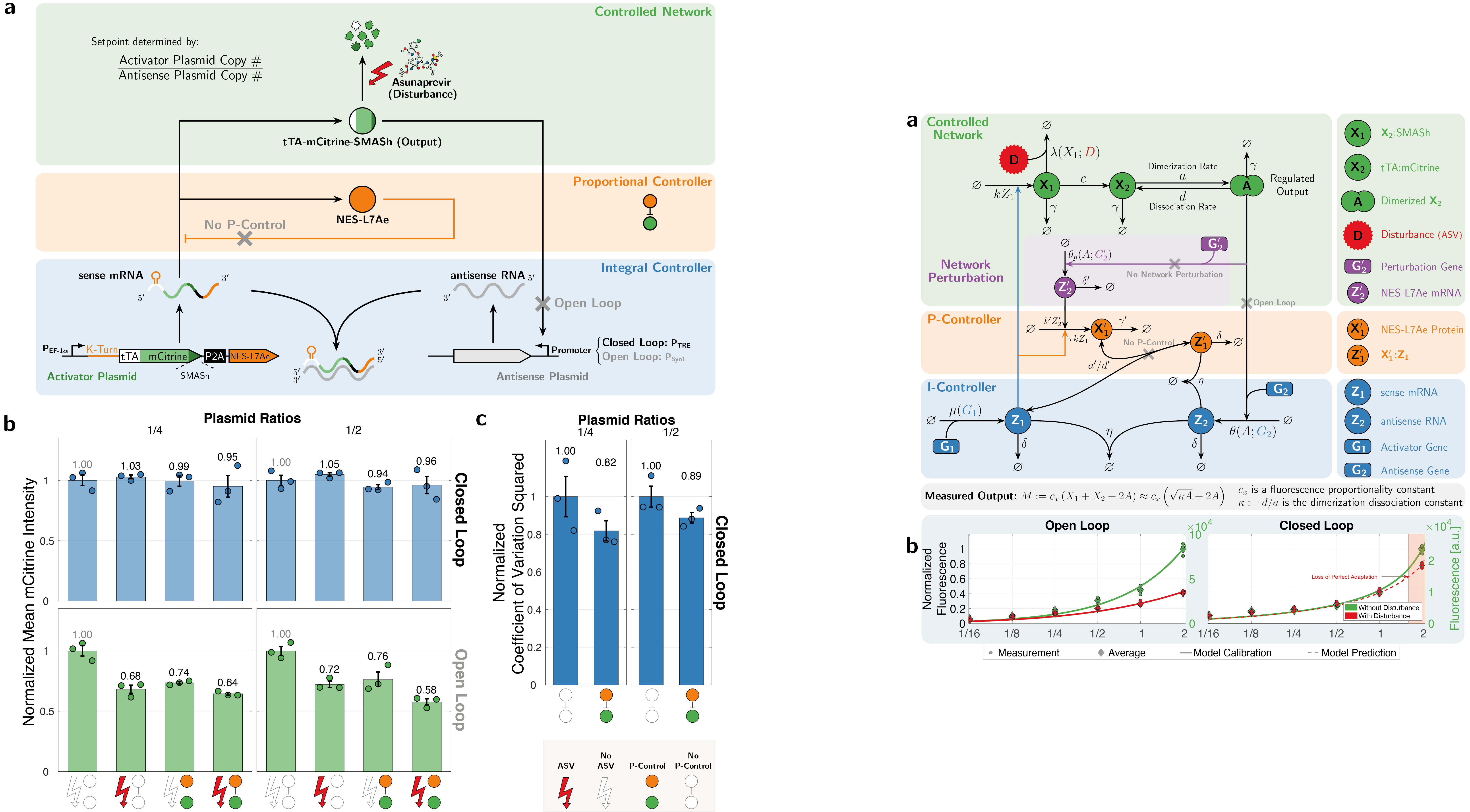
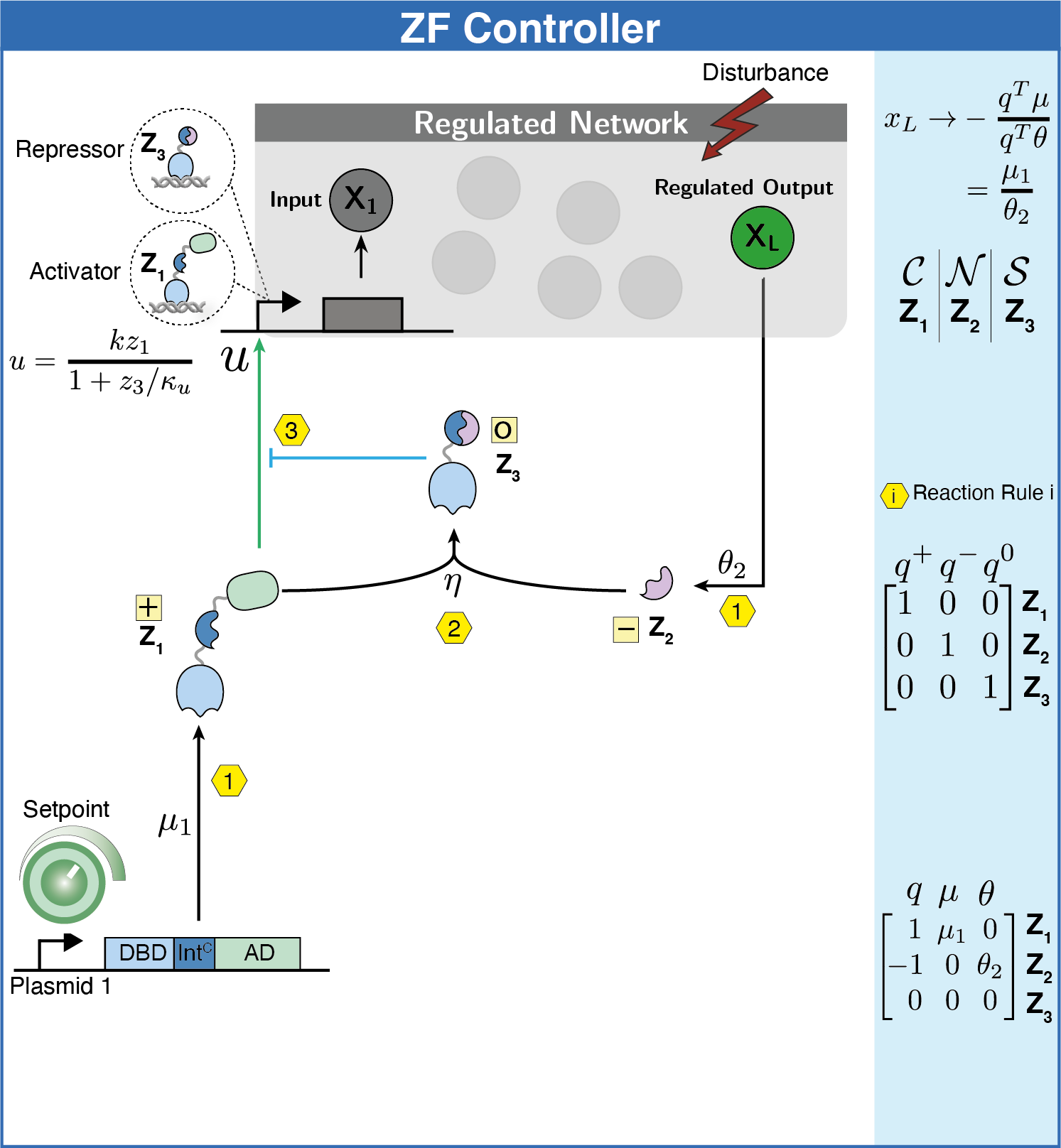
Subsequently, we explored protein-based implementations of these controllers in mammalian cells, which led us to inteins. Inteins are short protein segments that can autocatalytically cut themselves out of a protein structure while simultaneously rejoining the remaining pieces, known as exteins. A specific type, split inteins, consists of two separate parts, often called IntN and IntC. When activated, these split inteins can form a heterodimer and perform protein splicing reactions.
 |
 |
The flexibility of intein-based designs has opened up a new research avenue encompassing both theoretical and experimental efforts. Specifically, we have developed a couple of theorems that describe chemical reaction networks capable of achieving robust perfect adaptation with inteins or similar components. Additionally, we have introduced a model reduction recipe that allows for the mathematical analysis of complex genetic circuits. This theoretical approach has guided us in constructing a variety of genetic controllers, all of which have been experimentally demonstrated to achieve robust perfect adaptation.
 |
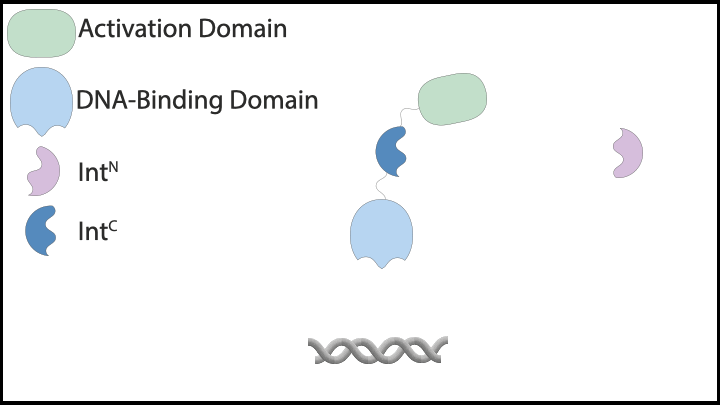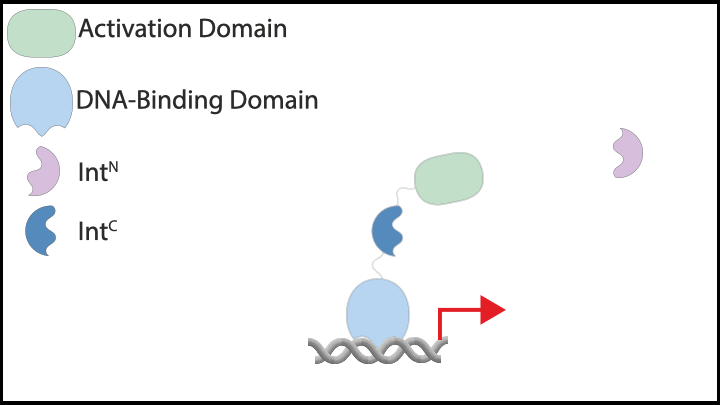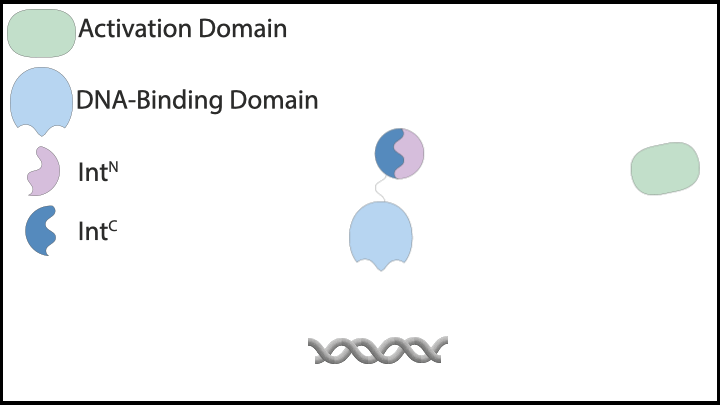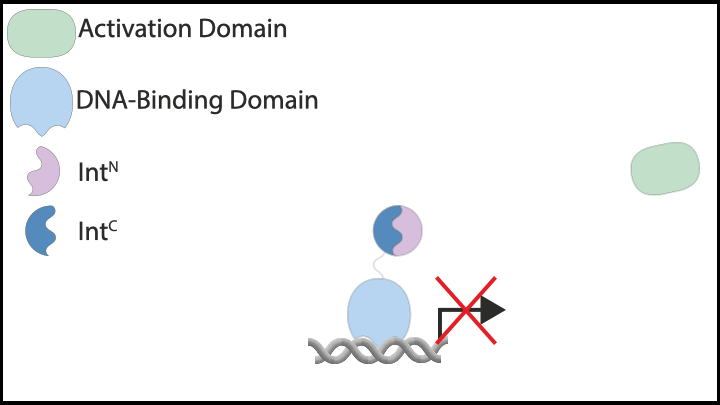 |
We value inteins greatly, so here’s a token of our appreciation:
The nonlinear dynamics in our controllers have yielded a surprising discovery. A design initially perceived as solely an integral controller was found to also incorporate a proportional controller "hidden" in the nonlinearities. This revelation is especially notable because both PI components are realized through a single actuation mechanism—a feature unprecedented in traditional control theory. This ultra-compact biomolecular PI controller has been successfully demonstrated to improve dynamic performance and reduce noise via simulations and analytical arguments.
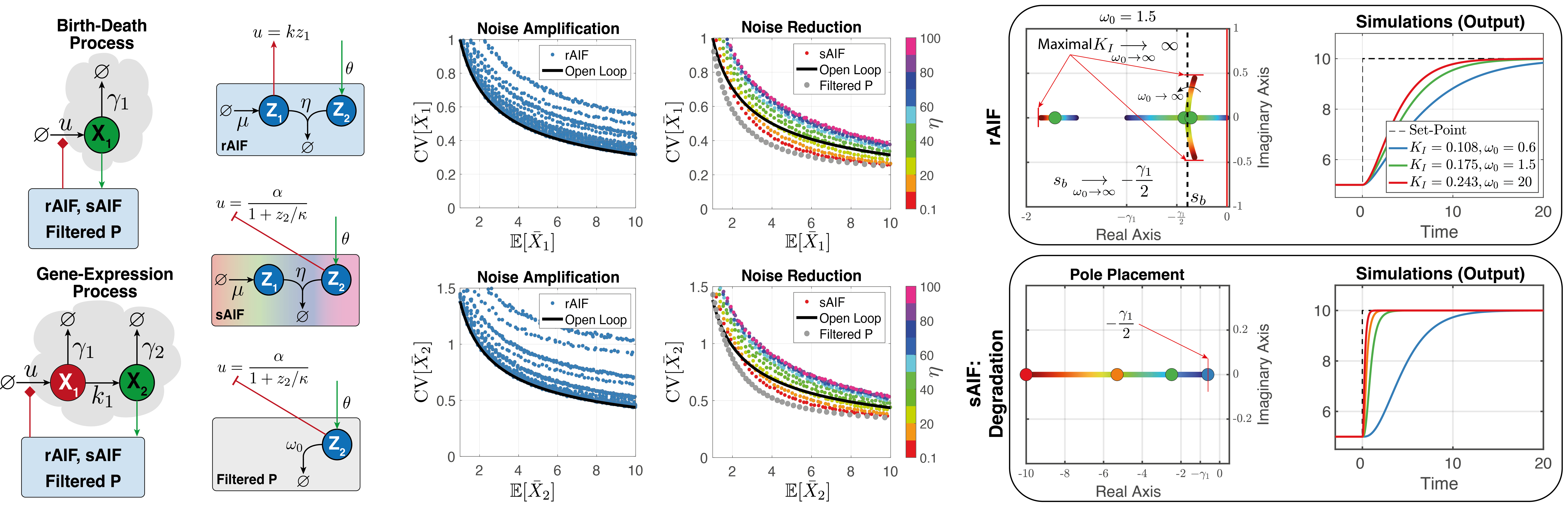
Motivated by our theoretical results, we implemented the controller in bacteria using inteins, and thus marking the first PI controller implementation in bacteria.
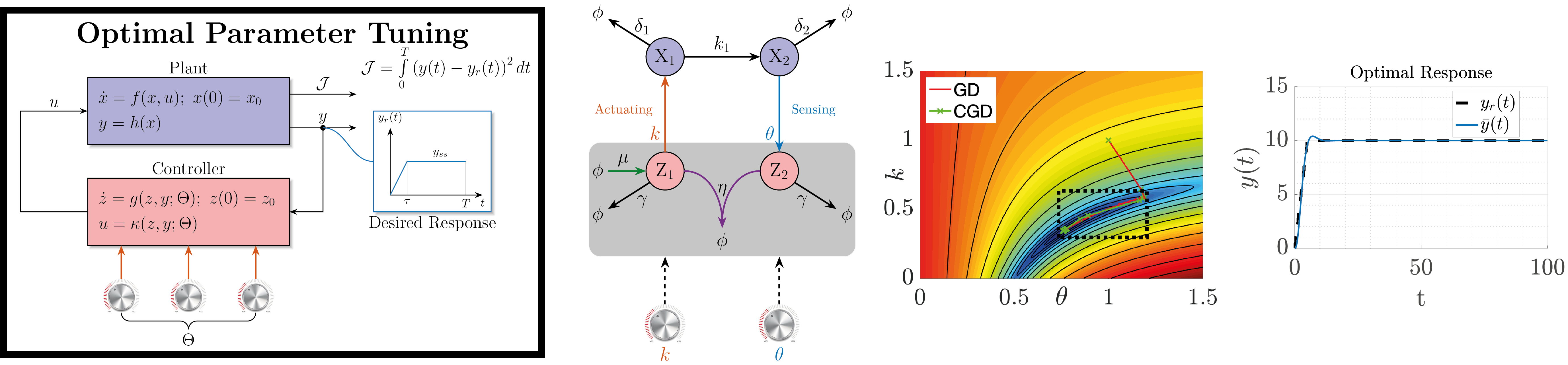
To maximize the effectiveness of feedback controllers, proper tuning is essential. To facilitate this, we have developed a framework and a numerical method similar to optimal control. This approach allows for the optimal tuning of nonlinear biomolecular feedback controllers, ensuring they meet specific objectives while adhering to predefined constraints.
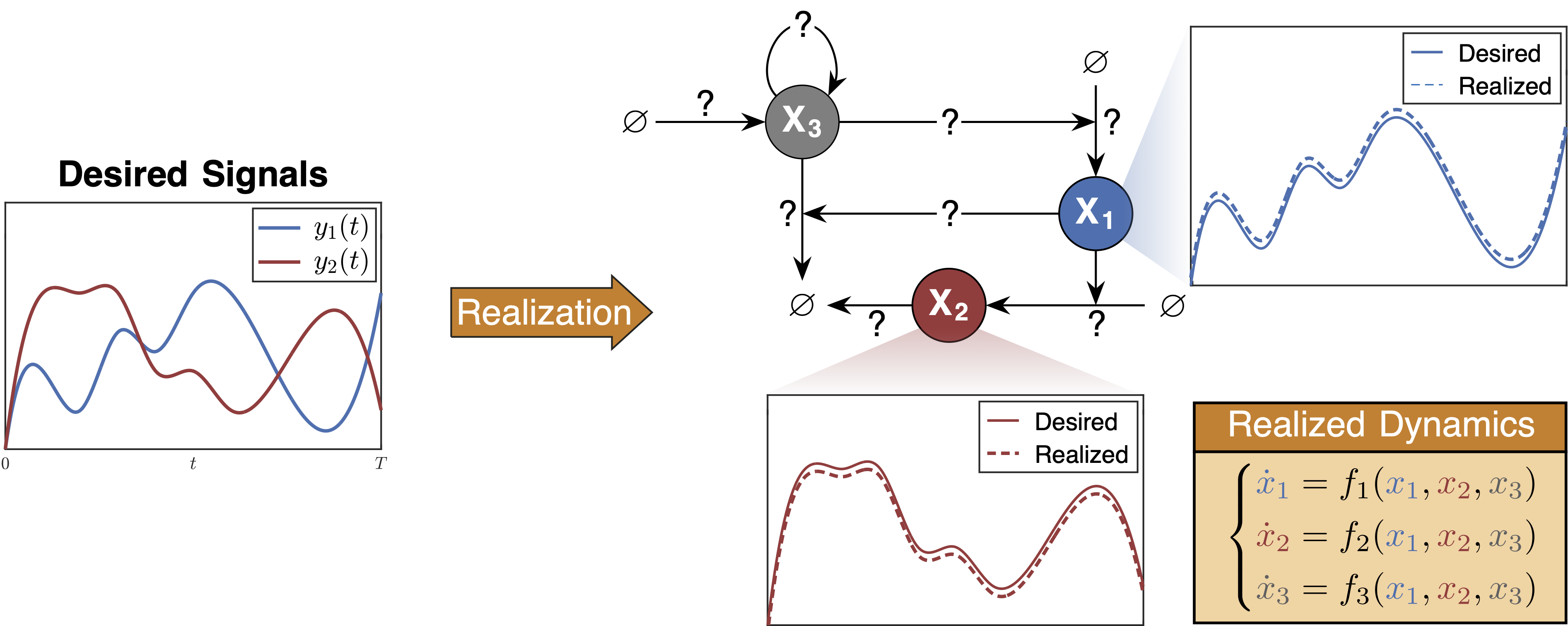
On the other hand, biomolecular systems are often characterized by a large number of molecular species and reactions, leading to complex models that are difficult to analyze and simulate. To address this, we have developed a method for reducing the complexity of biochemical reaction networks while preserving their essential dynamics. This approach simplifies the analysis using sparse and reduced biomolecular networks, making them more accessible for dynamical understanding.
References:
- Filo, Maurice, Sant Kumar, and Mustafa Khammash. "A hierarchy of biomolecular proportional-integral-derivative feedback controllers for robust perfect adaptation and dynamic performance." Nature communications 13, no. 1 (2022): 2119.
- Filo, Maurice, Ankit Gupta, and Mustafa Khammash. "Anti-windup strategies for biomolecular control systems facilitated by model reduction theory for sequestration networks." Science Advances 10, no. 34 (2024): eadl5439.
- Frei, Timothy, Ching-Hsiang Chang, Maurice Filo, Asterios Arampatzis, and Mustafa Khammash. "A genetic mammalian proportional–integral feedback control circuit for robust and precise gene regulation." Proceedings of the National Academy of Sciences 119, no. 00 (2022): e2122132119.
- Anastassov, Stanislav, Maurice Filo, Ching-Hsiang Chang, and Mustafa Khammash. "A cybergenetic framework for engineering intein-mediated integral feedback control systems." Nature Communications 14, no. 1 (2023): 1337.
- Anastassov, Stanislav, Maurice Filo, and Mustafa Khammash. "Inteins: A Swiss army knife for synthetic biology." Biotechnology Advances 73 (2024): 108349.
- Filo, Maurice, Ching-Hsiang Chang, and Mustafa Khammash. "Biomolecular feedback controllers: from theory to applications." Current Opinion in Biotechnology 79 (2023): 102882.
- Filo, Maurice, Mucun Hou, and M. Khammash. "A hidden proportional feedback mechanism underlies enhanced dynamic performance and noise rejection in sensor-based antithetic integral control." bioRxiv (2023): 2023-04.
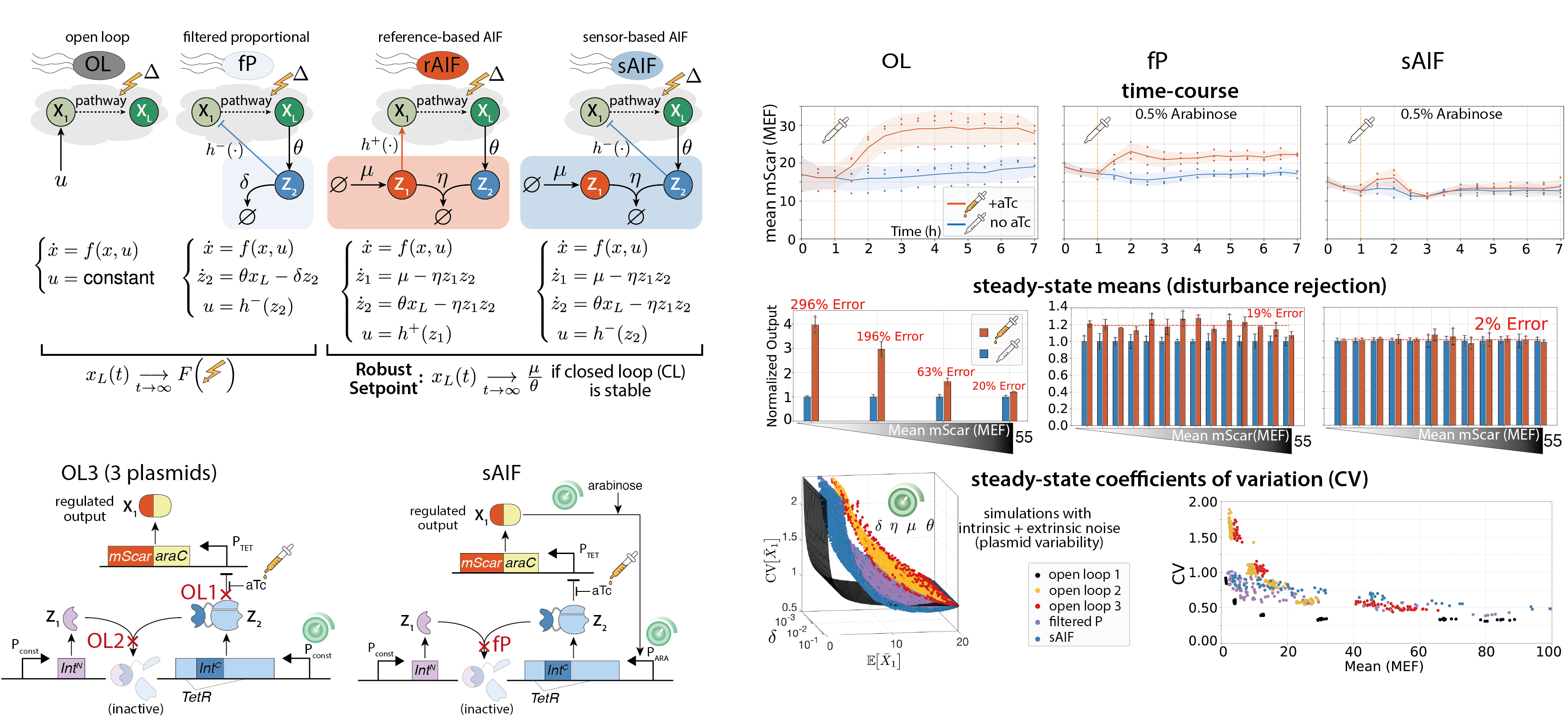
- Filo, Maurice, Stephanie Aoki, Mucun Hou, Stanislav Anastassov, and Mustafa Khammash. "Engineering Sensor-Based Antithetic Integral Controllers for Enhanced Dynamic Performance and Noise Attenuation," To appear in Cell Systems, 2026.
- Filo, Maurice, Sant Kumar, Stanislav Anastassov, and Mustafa Khammash. "Exploiting the nonlinear structure of the antithetic integral controller to enhance dynamic performance." 2022 IEEE 61st Conference on Decision and Control (CDC). IEEE, 2022.
- Filo, Maurice, and Mustafa Khammash. "Optimal parameter tuning of feedback controllers with application to biomolecular antithetic integral control." 2019 IEEE 58th Conference on Decision and Control (CDC). IEEE, 2019.
- Filo, Maurice, and Mustafa Khammash. "Realizing Reduced and Sparse Biochemical Reaction Networks from Dynamics." arXiv 2025.